Introduction. Although the
fundamental ideas of most experiments in biological science are
straightforward, the shear amount of data that has been produced is
mind boggling. It is beyond the ability (or the desire) of any of us
to recall or make use of all this information, yet when efficiently
used, these data provide a rich source of information. Luckily, the
advent of computer technology has provided the means for the
community to organize these data and make them available to the
community. The need to accomplish this task has created a new
discipline within the community of biological scientists to:
Specialists within the field of Bioinformatics work both independently and in collaboration with other scientists. Beyond the advances they make, the tools developed by this community can save you considerable time in your research. For example, in silica* searches are much faster than corresponding experimental approaches. An afternoon at the computer can often save weeks of laboratory work. Beyond speed, in silica approaches can reveal important evidence for evolutionary and functional relationships between genes and proteins.
The relationship of Bioinformatics to other pages on this site. This site is organized around the idea that current experimental approaches allow scientists to use information to bootstrap themselves from information about one type of molecule (DNA, RNA, protein, or antibody) to another type of macromolecule (see the home page).
Likewise, once this type of information is obtained and organized, links among different types of information allow the scientist to move among information about DNA, RNA, proteins, genetics, biological structures, and other information as is illustrated by the Entrez databases, which are under constant development by NCBI (National Center of Biotechnology Information) and collaborative scientists. Entrez's view of the relationship among databases and information is conceptualized by the following diagram, which was copied from their website (OMIM stands for On-Line Mendelian Inheritance in Man):
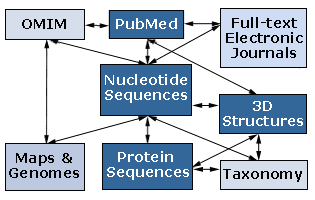
NCBI also has advanced workshops for bioinformatics and other online tutorials for many of their tools as well as tutorials in fundamental science concepts.
Then we will consider three additional ways that computer based approaches allow communication within the community and take advantages of commercial resources:
This page will also provide some links to resources, and we encourage users of this site to suggest additional links that may be of interest. There are several sites devoted to listing links to databases and computational research tools like Amos' WWW links page, the NCBI's Site Map, or CMS Molecular Biology Resources.
1. Bridging among protein, DNA, and RNA sequences. The central dogma predicts that the sequence of DNA predicts the sequence of RNA, which in turn determines the primary sequence of proteins.
Thus, partial sequence information from a gene of interest can be used to search for either corresponding cDNAs or genomic DNAs which may reside in a publicly accessible database. This can lead to clues about:
Likewise, partial sequence information from a protein can be used design a probe to screen a cDNA library, but it can also be used to query a nucleic acid data base for a protein with a related sequence.
Proteomic approaches produce a huge amount of data, and computational approaches can help manage that data. Algorithms can suggest the probable structure of protein fragments, including the complexity added by post-translational modifications as illustrated on the PROWL site or the links on the ExPASy Proteomics tools page. Once sequences are determined, the relationship of partial sequence data can be compared to database information to identify the corresponding protein, RNA, or gene.
2. Searching for related sequences in other organisms. Knowledge of sequence information in one organism can be used to search for corresponding genes in another organism. For example, if genetic information suggests that a particular gene is associated with a human disease, an in silica search (a search of databases using silicon based chips) can identify candidates for the corresponding gene in other species at either the protein or nucleotide level (see BLAST). Phenotypes observed in one organism, are at least indications of possible functions for the orthologous genes in other organisms. See, for example, NCBI's OMIM data base (Online Mendelian Inheritance in Man).
3. Searching for functional patterns in proteins and nucleic acids. Very often, the function or activity of an unknown protein can be ascertained by identifying relationships to known functional domains within its amino acid sequence. Computers provide a powerful way to identify specific patterns in sequence information.
4. Determine if there are known interactions among proteins. Frequently clues to the role of a protein can be developed by determining if a protein (or closely related proteins) are known to interact physically or interact indirectly as part of a known pathway. Databases, including known pathways and the proteins involved in those pathways, can can provide a rapid way of developing testable models for a proteins function. Some databases also collect and update information as more information on signaling pathways emerges. Some sites that can facilitate this include:
5. Structural studies and predictions. Web based computational programs and databases provide an accessible way of studying the structure and interactions of biological molecules. Programs can:
The ability to visualize and model molecular interactions is an invaluable approach to understanding biological processes and it is often an essential element in experimental design. The interplay between structural/energetic studies and functional tests of these model helps refine both approaches.
6. Managing data. In addition to sequence information, many current experimental approaches, including the measurement of gene expression or genotyping by DNA arrays, results in the accumulation of a large amount of data, which can become accessible only by incorporating it into a database. Likewise, comparison of data among labs or making data from one lab available to the scientific community requires sharing the information by web based programs. An excellent guide to managing gene expression data from microarrays is provided by Pat Brown's lab at Stanford, and the Stanford microarray database includes tools, databases and links to other resources. Weill is currently using the maxd system. ExPASy includes a page devoted to databases and analysis of 2D gels.
7. Getting in touch with the scientific literature. The scientific literature is rapidly being organized into a gigantic searchable relational database ( see Entrez databases). The scientific literature can be searched for keywords or combinations of keywords (genes, organisms, diseases, enzymatic activities, receptors, binding sites, enzymatic activities, metabolites, authors, etc). These searches can result in identifying primary references, single topic mini-reviews, and thorough, scholarly reviews. These papers are often linked to on-line databases. Tables of contents are often on-line and can be browsed. Many papers can be downloaded and/or printed. Understanding how to effectively use these search engines has become an essential scientific skill that can be developed by individual exploration of web sites or by organized classes taught by experts. Expert classes are available through many academic libraries, which are developing and sharing sophisticated computational and electronic resources. See PubMed - from NCBI; or see PubMed Central; or Medline from BioMedNet. The BioHUNT site at exPASy is another interesting search tool. A number of texts can be found on-line. PubMed has a library of books on line and Ergito provides text of Genes 2000.
8. Identifying reagents and protocols for their use from biotechnology companies. As cutting edge applications become more common, they are often commercialized. Likewise, these companies are carrying out independent research that often results in approaches that are useful for others in the community. Thus, the expertise of biotechnology companies and pharmaceutical companies becomes a valuable resource. These companies provide not only reagents, but also information about how the company believes the reagents can be used and scientific information about the fields where they can be used. To look up a company, try Hum-bolgen or Lab Velocity. Here are links to a few interesting sites:
9. Becoming aware of meetings where scientists exchange information and ideas. Meetings provide an opportunity of scientists to share their data, advertise their accomplishments, find collaborators, meet their competition, and enjoy being a member of a fast moving community. Meetings are organized by professional societies, educational institutions, independent corporations. Here are a few good sites:
10. Find funding opportunities for your ideas and find out what others are doing or want to do. Every institution has links to lists of funding sources. The Samuel J. Wood Library at the Weill Medical College has a section devoted to these sites, some of which are proprietary. A few of the more interesting include:
11. Learn so much about biomedical research that you can make a killing on the stock market.

See Club Biomed, an investment club and study group for students, fellows, faculty, and their friends.