Introductions and purpose. Not many years ago, those at the forefront of
molecular biology who had invented recombinant DNA technology,
sometimes like to call themselves genetic engineers. This term has
begun to fall out of favor, but it accurately describes the belief of
many individuals that the techniques and approaches that were being
developed by molecular biologists were sufficiently powerful to
design and modify life forms. As this enterprise has proceeded over
the last thirty years, biological scientists have repeatedly had to
appreciate the limitations of what they could accomplish. Yet, each
new limitation has inspired creative scientists to strain to overcome
those boundaries and those efforts have repeatedly been awarded with
powerful successes.
Currently we are in a position where the application of the techniques and logical approaches developed within biomedical sciences allow us to effectively approach the most central problems in biology and biomedical science. The youngest generation of emerging scientists has at its disposal a repertoire of tools that were unimaginable only a generation ago. The purpose of this site is to outline some of the major logical elements and technical approaches that are available and to show the relationship among these approaches. I hope that this overview will allow individuals to appreciate the logic that has been repeatedly applied by other scientists to attack fundamental biological problems and to get on with the most difficult and challenging enterprise that faces each young scientist:
Overview. The most powerful event in biological sciences in this century was the establishment that genes were encoded in DNA and the elucidation of the way the information encoded in those genes was expressed in biological systems. We now appreciate that the genetic material of most organisms is DNA and the predominant way that this information is expressed is by translation of DNA into RNA and the translation of RNA into proteins. Of course, there is much information in DNA that is not expressed as a messenger RNA. There are some genes that function purely by encoding RNAs that have a structural role in the cell. There are also cis-acting sequences in the DNA that allow it to be acted upon by specific DNA binding proteins that may regulate transcription of RNA, or even cause DNA rearrangements. There are also structural elements in DNA that are required for maintaining the structure of chromosomes. In some cases, RNA is reverse transcribed into DNA. Nevertheless, the basic idea of the central dogma *that:
is a useful simplification on which we can hinge the organization of the exploration of the logic of many biomedical studies.
 Biological
scientists constantly make use of the central dogma in understanding
biological problems; but, experimentally, they
use a variety of approaches to generate new information about each of
these polymers from existing knowledge of the others. Furthermore,
the use of antibodies has become such an important approach to
biological questions, that it deserves discussion. Beginning with
knowledge of protein structure, a cDNA sequence, a gene, or an
antibody, it is possible to get information about all of these
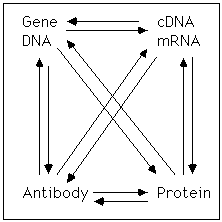
elements. The 'Experimental Dogma', which is illustrated in the
attached diagram, states that given any high quality reagent,
regardless of whether it is DNA, RNA, protein, or an antibody, it is
possible to boot-strap yourself to knowledge about the other elements
of the group. Furthermore, it is possible to begin with genetic
information (markers) and use it to isolate the gene or begin with a
cDNA or a genomic DNA and take a genetic approach to understanding
function. Often, comparison of new data to facts that have been
incorporated into databases produces important insights. This site
will help explain some of the ways these things can be done.
Biological
scientists constantly make use of the central dogma in understanding
biological problems; but, experimentally, they
use a variety of approaches to generate new information about each of
these polymers from existing knowledge of the others. Furthermore,
the use of antibodies has become such an important approach to
biological questions, that it deserves discussion. Beginning with
knowledge of protein structure, a cDNA sequence, a gene, or an
antibody, it is possible to get information about all of these
elements. The 'Experimental Dogma', which is illustrated in the
attached diagram, states that given any high quality reagent,
regardless of whether it is DNA, RNA, protein, or an antibody, it is
possible to boot-strap yourself to knowledge about the other elements
of the group. Furthermore, it is possible to begin with genetic
information (markers) and use it to isolate the gene or begin with a
cDNA or a genomic DNA and take a genetic approach to understanding
function. Often, comparison of new data to facts that have been
incorporated into databases produces important insights. This site
will help explain some of the ways these things can be done.
This site is not designed elucidate the details of any technique, rather it is designed to focus on the logic of experimental approaches. As the site evolves, it may provide links to other pages where technical questions and protocols are discussed.
Organization of the site. This work is divided into six pages (experimental design & logic (this page), proteins, mRNAs & cDNAs, antibodies, DNA & genes, and dictionary). Although ideas are developed in sequential fashion, it should also be possible read topics in any order. The dictionary of terms is being actively developed. It includes links to text where the topics are discussed, short explanations of the term, and links to pages where techniques or topics are explained in greater detail. Links that are followed with an asterisk (*) indicates a word is included in the dictionary, and clicking on the link will lead to the appropriate part of the dictionary to look up the work. (Sorry, but the program used to create this site (Claris Homepage) has some kind of bug or weakness that prevents providing links directly to all the dictionary entries. Claris has not yet been willing to provide any help or insight into this problem.)
Proteins. For convenience, it is easiest to begin our considerations with proteins, which will be the first part of this monograph. This section will consider the logic of protein purification and develop logical criteria that a particular biological activity resides in a particular protein. Of course, we will see that protein function can also be studied using cDNAs-based approaches and genomic approaches in other sections. This section also includes an long example that illustrates some basic principles in protein purification.
mRNAs & cDNAs. We will then consider how information about proteins can be used to identify and study specific messenger RNAs in the second section of this work. This discussion will be extended to a consideration of the ways understanding messenger RNA can provided a very powerful approach to many biological questions, including study of protein function and gene expression. We will also consider some of the other strategies that have allowed us to isolate messenger RNAs of interest.
Antibodies. In the third section, we will consider how scientists frequently take advantage of the ability of animals to make antibodies that recognize specific proteins to study protein expression and function.
DNA and genes. In this section, we will finally get to DNA which is the principle material which has been used to encode genes. We will consider how specific genes can be isolated from the genome and studied in isolation. This will lead us naturally to a consideration of the DNA sequences that are involved primarily in regulating gene expression and how these sequences can be isolated and studied. At the end of the section we will consider genes and how the logic implicit in genetic analysis provides a powerful tool to establishing the biological relevance of any protein, messenger RNA or DNA sequence that we might be studying. We will briefly review the tremendous advantages enjoyed by several established genetic systems, but focus primarily on how it is possible to manipulate genes in vertebrate organisms by approaches that either interrupt known sequences of DNA in an organism or add additional exogenous sequences of DNA (knock-out, knock-in, and transgenic approaches).
Interactions: Interactions between macromolecules are often biolgically important, so identifying these interactions is often a key problem. Some of the approaches discussed in other section are listed in the following table.
Interaction Detected Commonly Used Techniques
Protein-Protein Yeast Two-Hybrid Screen
Phage Display Library
Immunoprecipitation Assays
Protein-DNA DNA Footprinting
Gel Shift Analysis
Yeast One-Hybrid Screen
Southwestern Blot
Protein-RNA Yeast Three-Hybrid Screen
Bioinformatics: Computational tools that often draw on databases that are available through the internet provide an important link among DNA, RNA, and protein. These tools are used to build models that help refine our understanding of the ways macromolecules function and they help us see patterns in the structure of biological molecules and the metabolic pathways. they control.
Dictionary: A B C D E F G H I J K L M N O P Q R S T U V W X Y Z
There are two objectives that I hope this site will achieve:
First, there are extraordinarily important complimentary ideas among approaches that focus on the study of proteins, messenger RNAs, and genes.
Second, I hope you will appreciate the power of all of these approaches is simply not an adequate justification for their random application. There seem to be relatively few limits to what the next generation of biological researchers will be able to do with these powerful approaches, but there are more biological questions that can be asked than there are resources or time to accomplish. We can't do everything. Thus, the defining element of a scientist's quality will not be the ability to apply large numbers of technical approaches, but rather the ability to determine what biological questions should be addressed. What is most important and what is most likely to lead to a clear and interesting answer? This is not to say that development of additional techniques or technical approaches are not important. They are. Frequently our ability to get an answer depends on a technical breakthrough. The explosion of biological work in many areas can be traced to specific technical advances that are described below. But the value of any technical advance lies in whether it can be used effectively (and logically) to ask a biological question that is both important and interesting. One of your major challenges will be to participate in the scientific community's effort to establish what are most important and most interesting questions.
How to make additions to this site: Potential additions (including text (just pasted it
into the message) and diagrams (in PICT or GEF format as
attachments), thoughts, and suggestions should be sent to
jawagne@med.cornell.edu